
Introduction |
Design |
Help |
Tools |
FAQ |
Contact |
Publications |
| QTY/NTY Design for Protein Solubilizing |
|
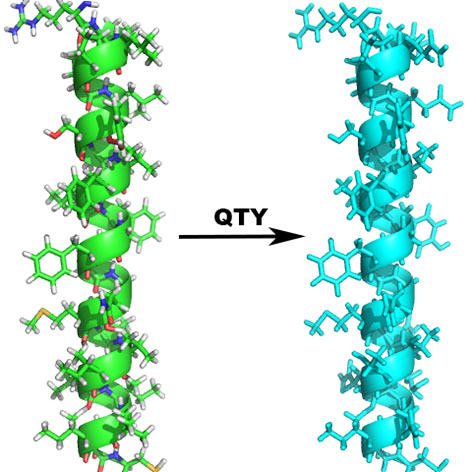
Membrane proteins play vital roles in all living systems. Approximately 30% of all genes in almost all sequenced genomes, code for membrane proteins. However, our detailed understanding of their structure and function lags far behind that of soluble proteins. As of September 2018, there are 144211 structures in the Protein Data Bank. However, there are only 1469 membrane protein structures with 825 unique structures including 28 G-protein coupled receptors (link). There are several bottlenecks in elucidating the structure and function of membrane receptors and their recognition and ligand-binding properties although they are of great interest. The most critical and challenging task is that it is extremely difficult to produce milligrams quantities of soluble and stable receptors. Inexpensive large-scale production methods are desperately needed, and have thus been the focus of extensive research. It is only possible to conduct detailed structural studies once these preliminary obstacles have been surmounted. In our just published work, with the GPCRs as model proteins, a code (QTY/NTY) based method was developed as an easy and efficient way for specially modifying the TM helixes and making the whole membrane protein water-soluble. The method has been used in making several GPCRs water-soluble including CCR5, CXCR4, CCR10 and CXCR7. This method is considered as a promising way which could be extensively applied for solubilizing alpha-helical proteins. Although the QTY/NTY code-based design is relative straightforward, the design job is still time-consuming if only a manual manner is available, especially for library design, which will be impractical without computing power. Here a web-based PSS sever was developed for facilitating the protein design based on the QTY/NTY code. This server is expected to be used widely by scientists who are endeavoring in the research about alpha-helical proteins. |
 |
 |
 |
 |
Copyright © 2013-2024 Shanghai Jiao Tong University
沪交ICP备20180143